rstudio drop in deviance test r|6.3.4 : Big box store We can use analysis of deviance tests (i.e., a likelihood ratio test of a nested model) to test the three-way interaction and the need for the . Resultado da 31 de mai. de 2023 · Seneca Buffalo Creek enforces a 12-team maximum on parlay cards at its self-service kiosks and a nine-team maximum at the betting counters in the sports lounges themselves. While a parlay card with more bets than this is extremely unlikely to pay out, limiting the maximum .
{plog:ftitle_list}
1 de dez. de 2023 · O chinês, que quase não teve tempo de colocar o short, tirou fotos da privada em chamas e compartilhou na rede social chinesa Weibo. Usuários ficaram chocados com a cena. Como explica o jornal chinês, a causa do incêndio não foi revelada, mas o proprietário suspeita que um curto-circuito tenha provocado as chamas.
I want to perform an analysis of deviance to test the significance of the interaction term. At first I did anova(mod1,mod2), and I used the function 1 - pchisq() to obtain a p-value .
In R, the drop1 command outputs something neat. These two commands .$Deviance = -2[L(\hat{\mathbf{\mu}} | \mathbf{y})-L(\mathbf{y}|\mathbf{y})]$ .In R, the drop1 command outputs something neat. These two commands should get you some output: example(step)#-> swiss. drop1(lm1, test="F") Mine looks like this: > drop1(lm1, . We can use analysis of deviance tests (i.e., a likelihood ratio test of a nested model) to test the three-way interaction and the need for the .
Adding S to the Null model drops the deviance by 36.41 − 0.16 = 36.25, and \(P(\chi^2_2 \geq 36.25)\approx 0\). So the S model fits significantly better than the Null model. And the S model fits the data very well.
regression
To fit a logistic regression model in R, use the glm function with the family argument set to binomial. To build a logistic regression model that predicts transmission using horsepower and miles per gallon, you can run the following . One commonly used method for doing this is known as leave-one-out cross-validation (LOOCV), which uses the following approach: 1. Split a dataset into a training set .Compute an analysis of deviance table for one or more generalized linear model fits. Usage ## S3 method for class 'glm' anova(object, ., dispersion = NULL, test = NULL)Compute all the single terms in the scope argument that can be added to or dropped from the model, fit those models and compute a table of the changes in fit.
The drop1() function compares all possible models that can be constructed by dropping a single model term. The add1() function compares all possible models that can be constructed by .$Deviance = -2[L(\hat{\mathbf{\mu}} | \mathbf{y})-L(\mathbf{y}|\mathbf{y})]$ However, most of the times, you want to test if you need to drop some variables. Say there are two models .In Section 4.4.4, the Wald-type test and drop-in-deviance test both suggest that a linear term in age is useful. But our exploratory data analysis in Section 4.4.2 suggests that a quadratic model might be more appropriate. A quadratic model would allow us to see if there exists an age where the number in the house is, on average, a maximum.
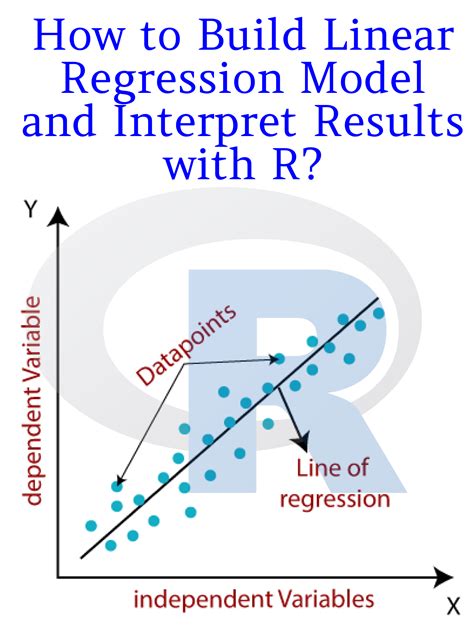
If you sum up the successes at each combination of the predictor variables, then the data becomes "grouped" or "aggregated". It looks like. X success failure n 0 1 0 1 1 0 2 2The difference between the null deviance and the residual deviance shows how our model is doing against the null model (a model with only the intercept). The wider this gap, the better. Analyzing the table we can see the drop in .
Step
Returns the deviance of a fitted model object. Rdocumentation. powered by. Learn R Programming. stats (version 3.6.2) Description. Usage Arguments . Value. Details. References. See Also .The table will optionally contain test statistics (and P values) comparing the reduction in deviance for the row to the residuals. For models with known dispersion (e.g., binomial and Poisson fits) the chi-squared test is most appropriate, and for those with dispersion estimated by moments (e.g., gaussian , quasibinomial and quasipoisson fits .
Step 2: Perform the Chi-Square Test of Independence. Next, we can perform the Chi-Square Test of Independence using the chisq.test() function: #Perform Chi-Square Test of Independence chisq.test(data) Pearson's Chi-squared test data: data X-squared = 0.86404, df = 2, p-value = 0.6492 The way to interpret the output is as follows:Analysis of deviance table. In R, we can test factors’ effects with the anova function to give an analysis of deviance table. We only include one factor in this model. So R dropped this factor (parentsmoke) and fit the intercept-only model to get the same statistics as above, i.e., the deviance \(G^2 = 29.121\). The following tutorials explain how to troubleshoot other common errors in R: How to Fix in R: names do not match previous names How to Fix in R: NAs Introduced by Coercion How to Fix in R: Subscript out of bounds How to Fix in R: contrasts can be applied only to factors with 2 or more levels $\begingroup$ Yes, an asymptotic chi-square result (if you use R, it's pchisq(2,1,lower.tail=FALSE)); it will correspond to a two-tailed z-test p-value (pnorm(sqrt(2),lower.tail=FALSE)*2), and so unless the d.f. are fairly small it will also closely approximate a t-test or F-test p-value cut off (above 40 d.f. it's 16% to the nearest whole .
water bottle brand ph test
3. Model assumptions. Count response: The response variable is a count (non-negative integers), i.e. the number of times an event occurs in an homogeneous time interval or a given space (e.g. the number of goal scored during a football game).It is suitable for grouped or ungrouped data since the sum of Poisson distributed observations is also Poisson. This increase in deviance is evidence of a significant lack of fit. We can also use the residual deviance to test whether the null hypothesis is true (i.e. Logistic regression model provides an adequate fit for the data). This is possible because the deviance is given by the chi-squared value at a certain degrees of freedom. The following example shows how to perform a likelihood ratio test in R. Example: Likelihood Ratio Test in R. The following code shows how to fit the following two regression models in R using data from the built-in mtcars dataset: Full model: mpg = β 0 + β 1 disp + β 2 carb + β 3 hp + β 4 cyl. Reduced model: mpg = β 0 + β 1 disp + β 2 carb Decision trees in R. Learn and use regression & classification algorithms for supervised learning in your data science project today! . which is similar to the Gini Index, is known as the Cross-Entropy or Deviance: The cross-entropy will take on a value near zero if the $\hat{\pi}_{mc}$’s are . This is the same as how we did with the test .
The test parameter can be set to "F" or "LRT". The default test is AIC. The direction parameter can be "both", "backward", or "forward". The following example does an F-test of the terms of the OLS model from above and a likelihood ratio test for several possible terms to the GLM model from above. Using drop1() and add1(). drop1 (mod, test = "F") ## chi square test p-value with (myprobit, pchisq (null.deviance - deviance, df.null - df.residual, lower.tail = FALSE)) ## [1] 7.219e-08 The chi-square of 41.56 with 5 degrees of freedom and an associated p-value of less .The "chi-square value" you're looking for is the deviance (-2*(log likelihood), at least up to an additive constant that doesn't matter for the purposes of inference. R gives you the log-likelihood above (logLik) and the likelihood ratio statistic (LR.stat): the LR stat is twice the difference in the log-likelihoods (2*(2236.0-2084.7)).
I wish to see the summary of deviance residuals when I run summary on a GLM in R. I believe this should be shown by default, however it doesn't appear for me. . ("deviance residuals", Text), db=db) This leads to the following. Changes in version 4.3.0 NEW FEATURES. The print() method for class "summary.glm" no longer shows summary statistics .Compute an analysis of deviance table for one or more generalized linear model fits. Rdocumentation. powered by. Learn R Programming. stats . test = "Cp") anova(glm.D93, test = "Chisq") glm.D93a <- update(glm.D93, ~treatment*outcome) # equivalent to Pearson Chi-square anova(glm.D93, glm.D93a, test = "Rao") # }
1 - (Residual Deviance/Null Deviance) If you think about it, you're trying to measure the ratio of the deviance in your model to the null; how much better your model is (residual deviance) than just the intercept (null deviance). If that ratio is tiny, you're 'explaining' most of the deviance in the null; 1 minus that gets you your R-squared. The following example shows how to perform a Wald test in R. Example: Wald Test in R. For this example, we’ll use the built-in mtcars dataset in R to fit the following multiple linear regression model: mpg = β 0 + β 1 disp + β 2 carb + β 3 hp + β 4 cyl. The following code shows how to fit this regression model and view the model summary: At the cost of 0.97695 extra degrees of freedom, the residual deviance is decreased by 1180.2. This reduction in deviance (or conversely, increase in deviance explained), at the cost of <1 degree of freedom, is highly unlikely if the true effect of x2 were 0. Why 0.97695 degrees of freedom increase?5.5 Deviance. The deviance is a key concept in generalized linear models. Intuitively, it measures the deviance of the fitted generalized linear model with respect to a perfect model for the sample \(\{(\mathbf{x}_i,Y_i)\}_{i=1}^n.\) This perfect model, known as the saturated model, is the model that perfectly fits the data, in the sense that the fitted responses (\(\hat Y_i\)) equal .
Introduction. ANOVA (ANalysis Of VAriance) is a statistical test to determine whether two or more population means are different. In other words, it is used to compare two or more groups to see if they are significantly different.. In practice, however, the: Student t-test is used to compare 2 groups;; ANOVA generalizes the t-test beyond 2 groups, so it is used to .Scaled deviance, defined as D = 2 * (log-likelihood of saturated model minus log-likelihood of fitted model), is often used as a measure of goodness-of-fit in GLM models. Percent deviance explained, defined as [D(null model) - D(fitted model)] / D(null model), is also sometimes used as the GLM analog to linear regression's R-squared.Clear examples in R. Choosing a statistical test. Summary and Analysis of Extension Program Evaluation in R. Salvatore S. Mangiafico. Search Rcompanion.org . Contents . Introduction . Quantile Regression Analysis of Deviance Table Df Resid Df F value Pr(>F) 1 1 200 5.7342 0.01756 * References [IDRE] Institute for Digital Research and .
R: Analysis of Deviance for Generalized Linear Model Fits
The \chi^2 test can be an exact test (lm models with known scale) or a likelihood-ratio test or a test of the reduction in scaled deviance depending on the method. For glm fits, you can also choose "LRT" and "Rao" for likelihood ratio tests and Rao's efficient score test. You can use the following syntax to calculate the standard deviation of a vector in R: sd(x) . Prev Dixon’s Q Test: Definition + Example. Next Lurking Variables: Definition & Examples. Leave a Reply Cancel reply. Your email address will .
Free account Reset your account password: If you reset your .
rstudio drop in deviance test r|6.3.4